
Research
Our research is focused on understanding fundamental structural, biochemical, and functional attributes for quality control pathways involving post-translational protein modification by ubiquitin and ubiquitin-like proteins such as SUMO, and those that contribute to co- and post-transcriptional RNA maturation, processing and decay. We pursue biochemical approaches to reconstitute key intermediates or complexes in these pathways and combine them with single particle cryo-EM and x-ray crystallography in conjunction with genetics and biochemistry to elucidate structure/activity relationships.
Ubiquitin and ubiquitin-like conjugation cascades regulate nearly every facet of eukaryotic biology either by targeting proteins for degradation or by generating new molecules that contribute to signal transduction. Ubiquitin and ubiquitin-like proteins are conjugated to their substrates through a series of reactions catalyzed by conserved enzymes and factors. Most of our early work focused on the Ubl protein SUMO as it is essential in budding yeast and its conjugation to substrates requires an enzyme cascade that involves fewer factors in comparison to the ubiquitin conjugation cascade thus facilitating both structural and functional studies. Our current efforts are focused on underst encompass both SUMO and ubiquitin in studies that address their activation, conjugation and recognition.
Figure 1
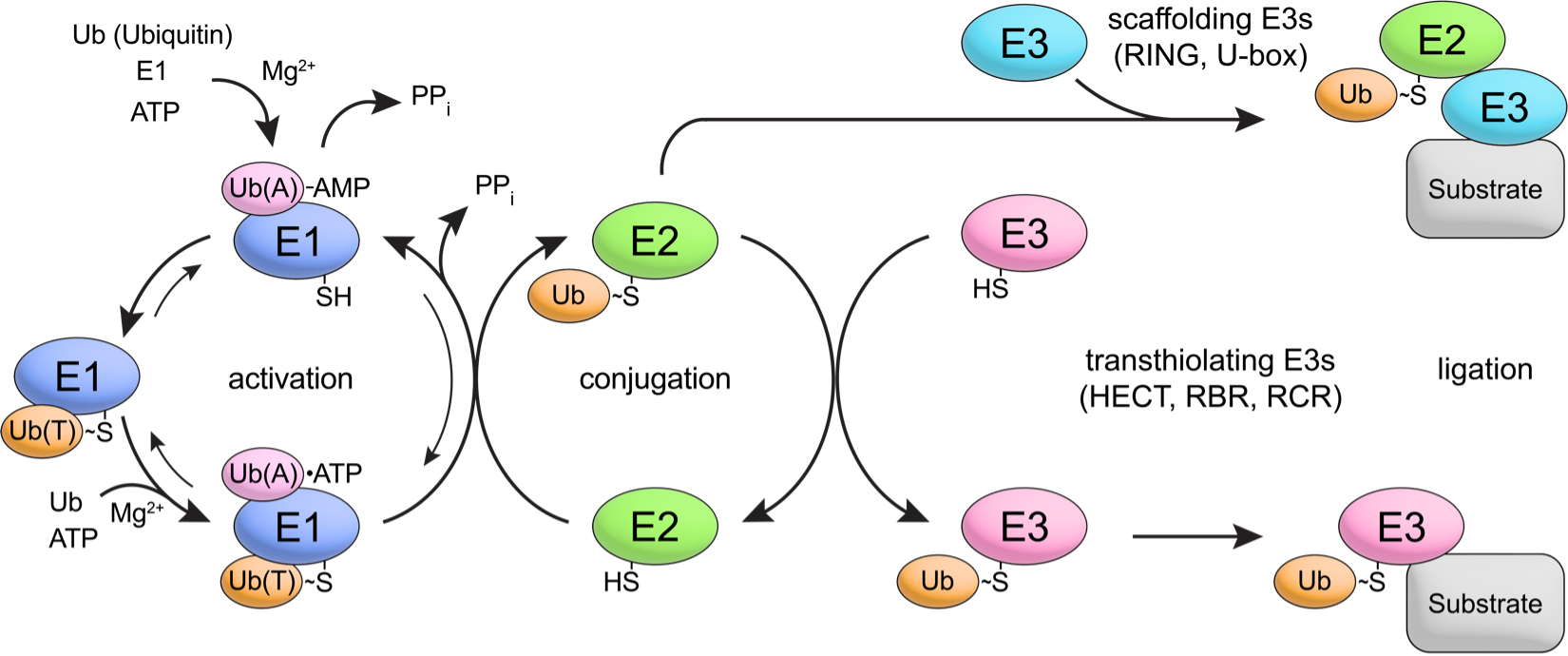
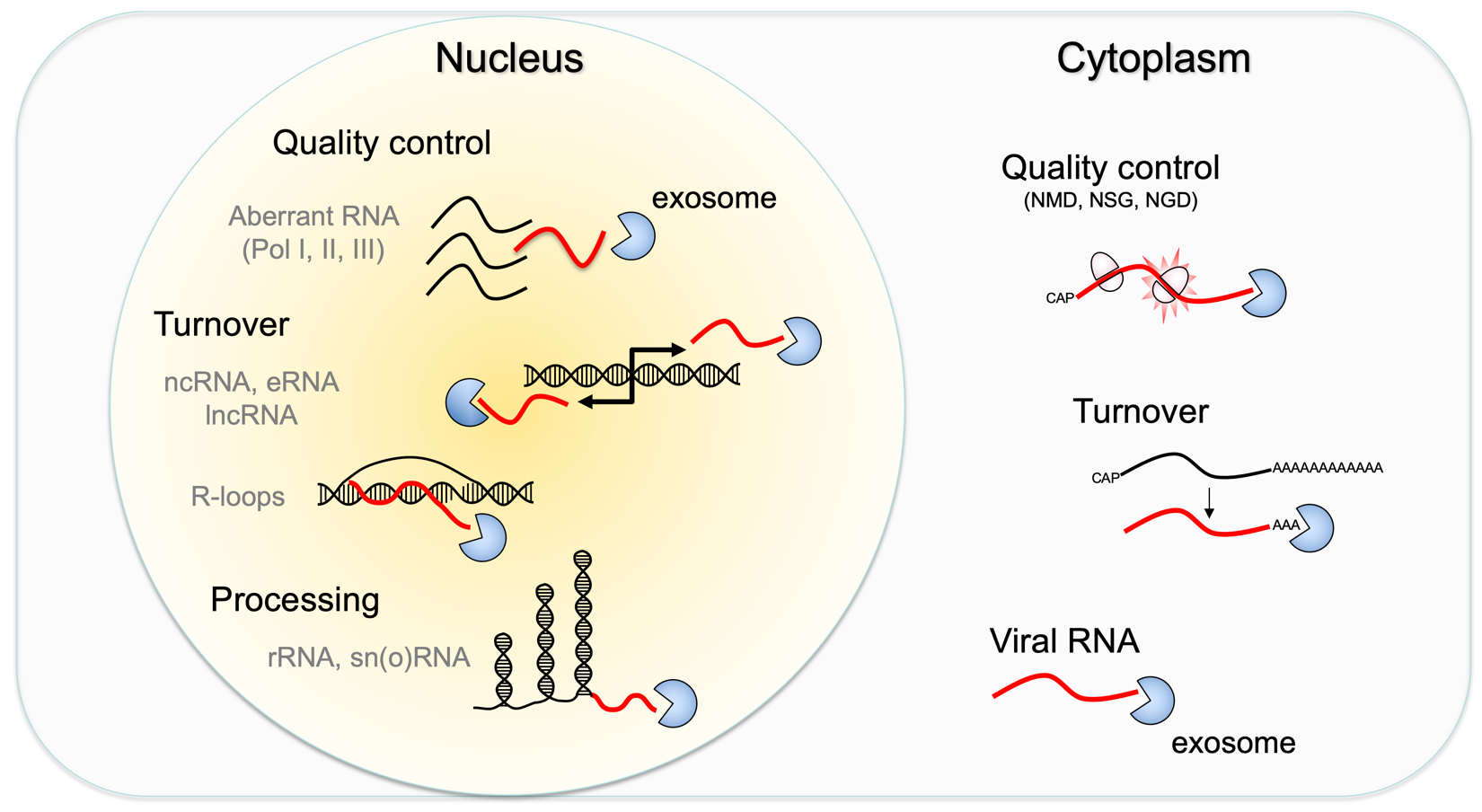
Another major area of investigation involves pathways that contribute to RNA quality control. RNA abundance is regulated by balancing transcription and degradation in processes that control the temporal and spatial distribution of cellular RNA. In eukaryotic cells, mRNA decay is catalyzed by two major pathways, and both can be initiated by deadenylation of the polyadenylated (poly-A) tail. After decapping, 5’ to 3’ RNA degradation is accomplished by Xrn1, a 5’ to 3’ exoribonuclease. In the 3’ to 5’ pathway, RNA degradation is catalyzed by a multi-subunit 3’ to 5’ exoribonuclease complex, the RNA exosome. The exosome also participates in processing small nucleolar and small nuclear RNAs (snoRNAs, snRNAs) and ribosomal RNAs (rRNAs) and degradation of many non-coding RNAs including those that arise from bidirectional transcription or transcription from enhancers.
Figure 2
Current Projects:
- Structure/function analysis of Ubiquitin and Ubiquitin-like conjugation cascades
- Structure/function analysis of factors involved in RNA decay and quality control
Bio
Christopher D. Lima is a Howard Hughes Medical Institute Investigator at the Sloan Kettering Institute where he is Chair and Member of the Structural Biology Program. He holds an Alfred P. Sloan Chair and is a Professor in the Weill Cornell and Sloan Kettering Graduate Schools. He received his PhD from Northwestern University for work on DNA topoisomerase I. As a Helen Hay Whitney Fellow at Columbia University, his work focused on resolving mechanisms underlying nucleotidyl transferases. He joined the faculty at Weill Cornell in 1998 and moved to Sloan Kettering in 2003. His research investigates RNA and protein quality control pathways. He was a Beckman Young Investigator and Rita Allen Scholar and was selected as an HHMI Investigator in 2013. He was elected to the American Academy of Arts and Sciences in 2017 and the National Academy of Sciences in 2020.
Distinctions:
- National Academy of Sciences
- American Academy of Arts and Sciences
- Howard Hughes Medical Institute
- Rita Allen Scholar
- Beckman Young Investigator
