
Research
Epigenetic regulation of gene expression is tied to numerous processes that determine cell phenotype and fate. Abrogation of epigenetic cascades drives various disease states, including cancer. Much of the work delineating the underlying mechanisms behind epigenetic control has contributed to the notion that a so-called ‘histone code,’ which refers to the landscape of histone enzymatically installed post-translational modifications (PTMs), is a key determinant of a gene's activity and its potential to be activated or repressed in response to environmental stimuli. Although increasing evidence has indicated the potential significance of histone PTMs in genomic regulation, we are still in the earliest stages of examining the many chromatin states linked to mammalian transcription and cellular response. Furthermore, much debate still exists as to whether histone PTMs are causally responsible for differences in chromatin states, and thus gene expression, or whether such differences are simply the consequences of dynamic processes associated with transcription and/or chromatin remodeling.
The highly interdisciplinary and innovative work in my lab aims to implement chemistry in biological research to address key questions in the epigenetics field by (1) developing and applying powerful chemical tools to investigate the mechanisms that drive epigenetically regulated transcription and (2) characterize chemical modifications on chromatin that vitiate its function and ultimately lead to disease. My lab developed novel in vitro and in vivo methodologies, including chemical probes and the first ever synthesis of site-specifically modified histones in animal brain. We applied these and other chemical-biology methods to address important questions in the field and make biological discoveries as well as reveal new therapeutic avenues in cancer. In one of our leading projects, we identified a new family of histone modifications that alter chromatin structure and function and serve as a new chapter in the epigenetic code. Identifying this new pathway in cancer provides a paradigm shift in the field of chromatin biology and opens a door to new cancer therapeutic targets. Indeed, we identified new key enzymatic regulators of this cascade, to which we are developing small molecule inhibitors.
Figure 1.
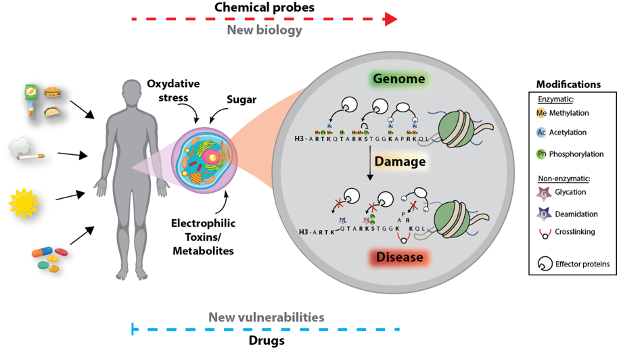

Current Projects:
- Non-enzymatic covalent modifications as a new link between metabolism and cell fate
- Characterizing the role of H1 linker histone in shaping the epigenetic landscape
- Identifying and characterizing new histone ubiquitination events
- Investigating the role of epigenetic regulation in HBV infection
- Studying non-canonical chromatin structures and their epigenetic landscape (e.g., micronuclei)
Bio
Dr. Yael David received her B.S. in Biology from SUNY Stony Brook, as a Summa Cum Laude after which she moved to the Weizmann Institute of Science in Israel, where was trained as a biochemist and cell biologist, applying her knowledge to study the mechanism and regulation of polyubiquitination. Realizing the power of interdisciplinary research, she moved to the Chemistry Department at Princeton University where she combined her experience with Prof. Tom Muir’s expertise in peptide chemistry to develop novel tools towards the mechanistic investigation of histone post-translational modifications including their site-specific manipulation in live cells. In 2016, Yael brought her powerful program to the Chemical Biology Program at Memorial Sloan Kettering Cancer Center.
Distinctions:
- Boyer Award in Basic Research
- ChemBio Talents, ChemBioChem
- STARR Cancer Consortium award
- NIH/NIGMS Maximizing Investigators' Research Award (MIRA) (R35)
- Anna Fuller Cancer Research Award
- Pershing Square Sohn Cancer Alliance (PSSCA) Award
- Chemical Protein Synthesis (CPS) “Rising Star in Protein Synthesis”
- American Chemical Society (ACS), “Future of Biochemistry”
- NIH Cutting-Edge Basic Research Award (CEBRA) Award (R21)
