
Research
Parasites that infect and live in human red blood cells have had to adapt in order to live in a hostile host environment. We study the vector-borne diseases, malaria and babesiosis, that have a large impact on human health. The parasites that cause these diseases have adapted differently to life in a red blood cell. Both, however, cause significant disease and malaria, in particular, is associated with a high morbidity and mortality especially in Sub-Saharan Africa. Besides causing acute symptoms, both parasites have found ways to persist in their host. We study processes that allow these parasites to cause disease, avoid host immune clearance and look for new therapeutic targets.
How does the parasite generate a diverse set of surface antigens? Malaria parasites have clusters of diverse multicopy gene families known to be important for virulence and maintaining a chronic infection. We study how the parasite maintains diversity in these gene families while preserving the functionality of the parasite core genome. We propose that the haploid nature of the parasite genome, the unique set of DNA repair pathways that are present and active in the parasite, and genome organization all play important roles in generating diversity in specific gene clusters.
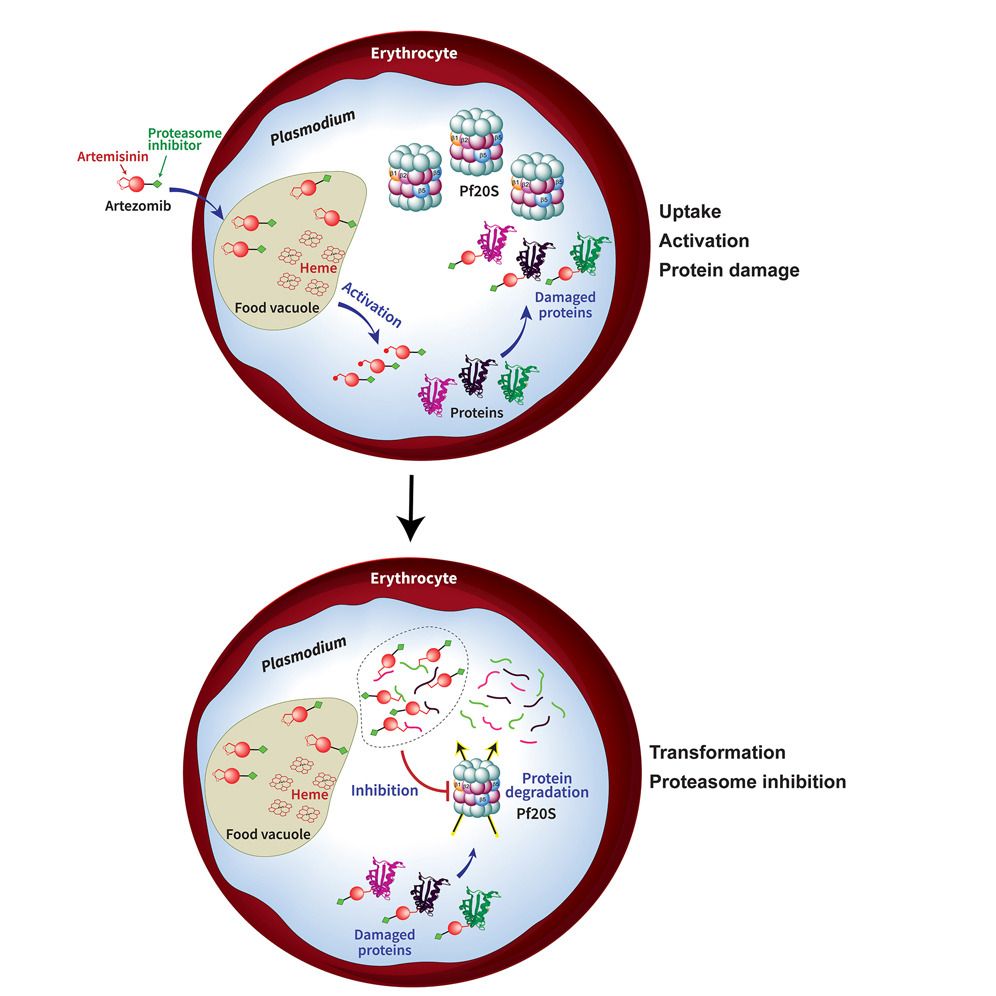
Is the parasite proteasome an effective antimalarial drug target? We work closely with the Lin lab in Microbiology and Immunology to develop and test parasite selective proteasome inhibitors as novel antimalarials. We also investigate how the parasite may develop resistance to proteasome inhibitors and the role the proteasome plays in parasite survival.
How does the babesia parasite avoid clearance in the human host and how can we best interrupt that process? Our translational research focuses on the parasitic tickborne disease prevalent in the Northeast, babesiosis. We are working to identify what makes different hosts experience more severe disease, what treatment combinations are the most effective and how can we best prevent relapsing complicated disease.
Parasites that infect and live in human red blood cells have had to adapt in order to live in a hostile host environment. We study the vector-borne diseases, malaria and babesiosis, that have a large impact on human health. The parasites that cause these diseases have adapted differently to life in a red blood cell. Both, however, cause significant disease and malaria, in particular, is associated with a high morbidity and mortality especially in Sub-Saharan Africa. Besides causing acute symptoms, both parasites have found ways to persist in their host. We study processes that allow these parasites to cause disease, avoid host immune clearance and look for new therapeutic targets.
How does the parasite generate a diverse set of surface antigens? Malaria parasites have clusters of diverse multicopy gene families known to be important for virulence and maintaining a chronic infection. We study how the parasite maintains diversity in these gene families while preserving the functionality of the parasite core genome. We propose that the haploid nature of the parasite genome, the unique set of DNA repair pathways that are present and active in the parasite, and genome organization all play important roles in generating diversity in specific gene clusters.
Is the parasite proteasome an effective antimalarial drug target? We work closely with the Lin lab in Microbiology and Immunology to develop and test parasite selective proteasome inhibitors as novel antimalarials. We also investigate how the parasite may develop resistance to proteasome inhibitors and the role the proteasome plays in parasite survival.
How does the babesia parasite avoid clearance in the human host and how can we best interrupt that process? Our translational research focuses on the parasitic tickborne disease prevalent in the Northeast, babesiosis. We are working to identify what makes different hosts experience more severe disease, what treatment combinations are the most effective and how can we best prevent relapsing complicated disease.
Figure 1 Babesia and Malaria
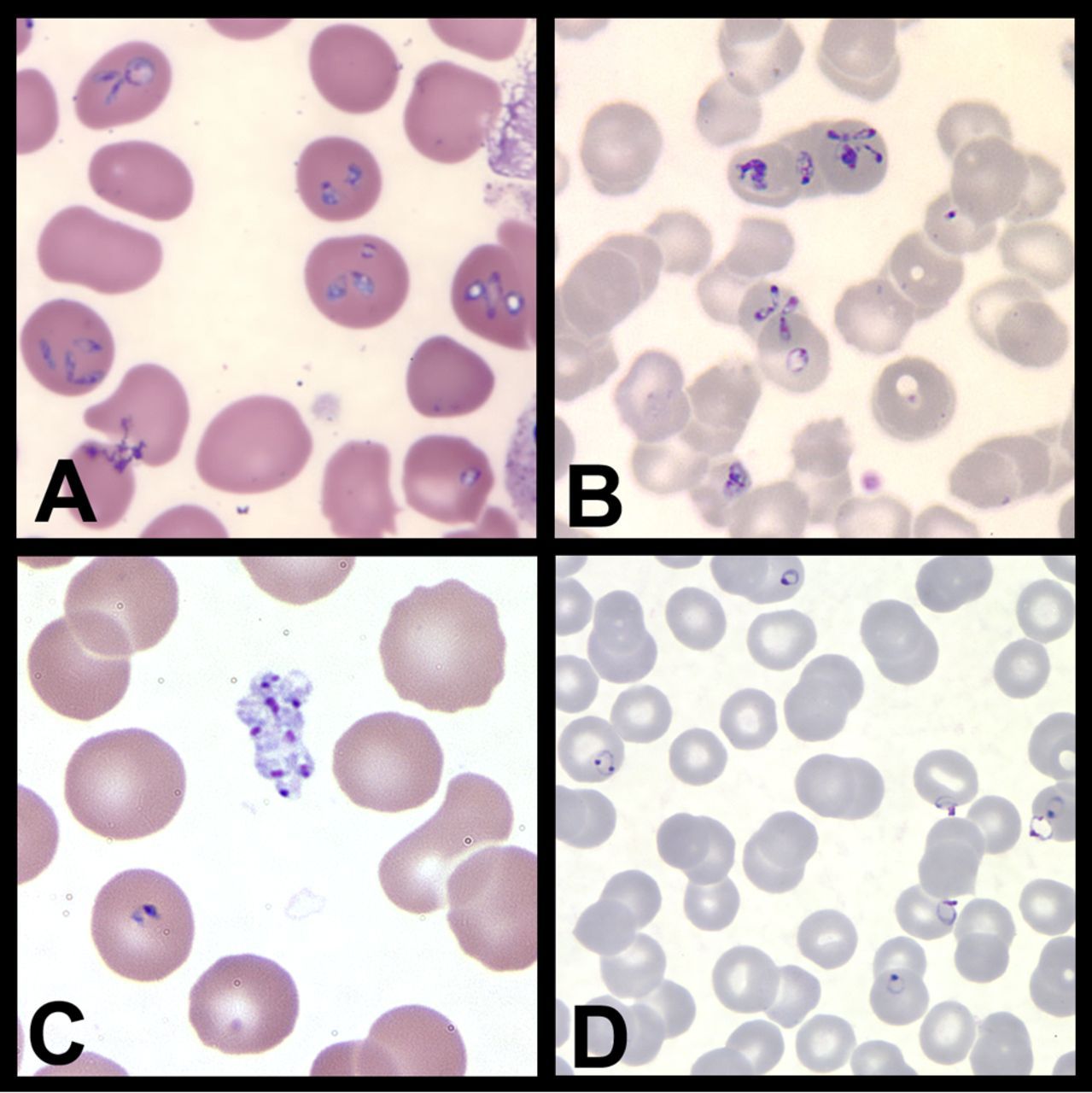
Figure 2 Artezomib
Current Projects:
- Genome maintenance in Plasmodium falciparum - We are working to define DNA repair pathways important to genome maintenance in the malaria parasite.
- The Parasite Proteasome as a Drug Target- We are using chemical and biological approaches to study the parasite ubiquitin-proteasome system as a potential target for antimalarial interventions.
- Understanding Human Babesiosis - We are interested in studying how babesia can persist in the host and which potential therapies would be most effective and lead to parasite clearance even in the immunosuppressed.
Bio
Dr. Laura Kirkman is an Associate Professor of Medicine and Microbiology and Immunology at Weill Cornell Medicine. Dr. Kirkman is a physician-scientist whose research focuses on the molecular pathogenesis of infection with bloodborne parasitic diseases: malaria and babesiosis. Dr. Kirkman received her M.D. from the Albert Einstein College of Medicine with distinction in research where she benefitted from support from a Howard Hughes Medical Student research award. She completed her clinical training in Internal Medicine at Yale-New Haven Hospital and her Infectious Disease fellowship at the New York-Presbyterian-Weill Cornell Medical Center.
