
Research
Question
What underlies diverse human traits? Why are some of us at a higher risk of developing certain diseases? How does genetic and epigenetic mechanisms regulate cell fate decisions in human development?
Approach
The Huangfu lab seeks to decode complex human disease trait through understanding the roles of genes and variants and their interactions with the environment. Our primary focus revolves around pancreas development and diabetes – a pressing global health issue. We use human pluripotent stem cells (hPSCs), including hESCs and hiPSCs, to recapitulate pancreas development and diabetes-relevant cell types. Our approach is multi-faceted, employing a range of strategies that encompass natural genetic perturbations (examining genetic variants in noncoding regions inherent to individuals' genetic makeup), deliberate experimental genetic perturbations (inclusive of CRISPR-Cas9 editing, CRISPR interference, and CRISPR activation), and exposure to stress-mimicking environmental conditions. To characterize the impact of these diverse perturbations, we conduct a variety phenotypic assay including scRNA-seq and Perturb-seq (combining CRISPR perturbation with scRNA-seq). These approaches have led us to discover many new genes and enhancer elements involved in the regulation of human pancreas biology, as well as fundamental epigenetic regulatory mechanisms (e.g., DNA methylation). In future studies, we aim to further deepen the understanding of the genes, variants, and environmental factors that contribute to cellular phenotypes and affect diabetes risks, thus informing the development of new therapies and preventive measures.
Figure 1
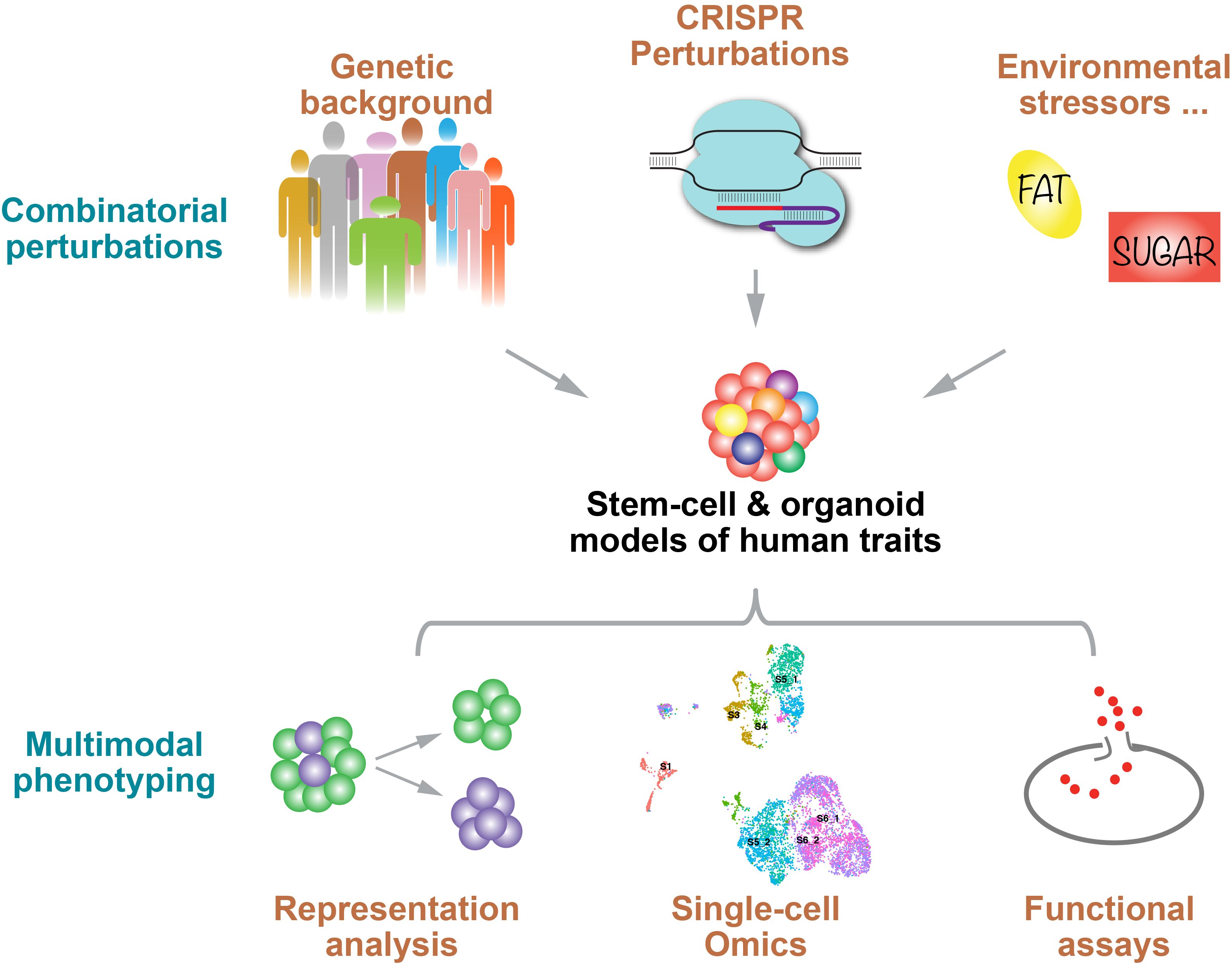
Current Projects:
- Genetic screens via CRISPR
- Enhancer discovery
- DNA methylation
- Perturb-seq
Bio
Huangfu earned her bachelor’s degree in Genetics from Fudan University (Shanghai, China), and her Ph.D. degree in Neuroscience from Cornell University Weill Graduate School of Medical Sciences in 2005. Her Ph.D. work with Kathryn V. Anderson uncovered a previously unsuspected role of primary cilia, a microtubule-based cellular organelle, in Hedgehog signal transduction and spinal cord development in mammals. She subsequently developed improved methods for the generation of iPSCs through reprogramming as a postdoctoral researcher with Douglas A. Melton at Harvard University. In 2010, she joined Sloan Kettering Institute and WCGS, and is now a Member and Professor.
Distinctions:
- Helen Hay Whitney Postdoctoral Fellowship (2006)
- Discovered the role of cilium in Hedgehog signal transduction
- Large-scale CRISPR screening in hPSCs
- Leading MorPhiC efforts to define null allele phenotypes in human cells
