
Research
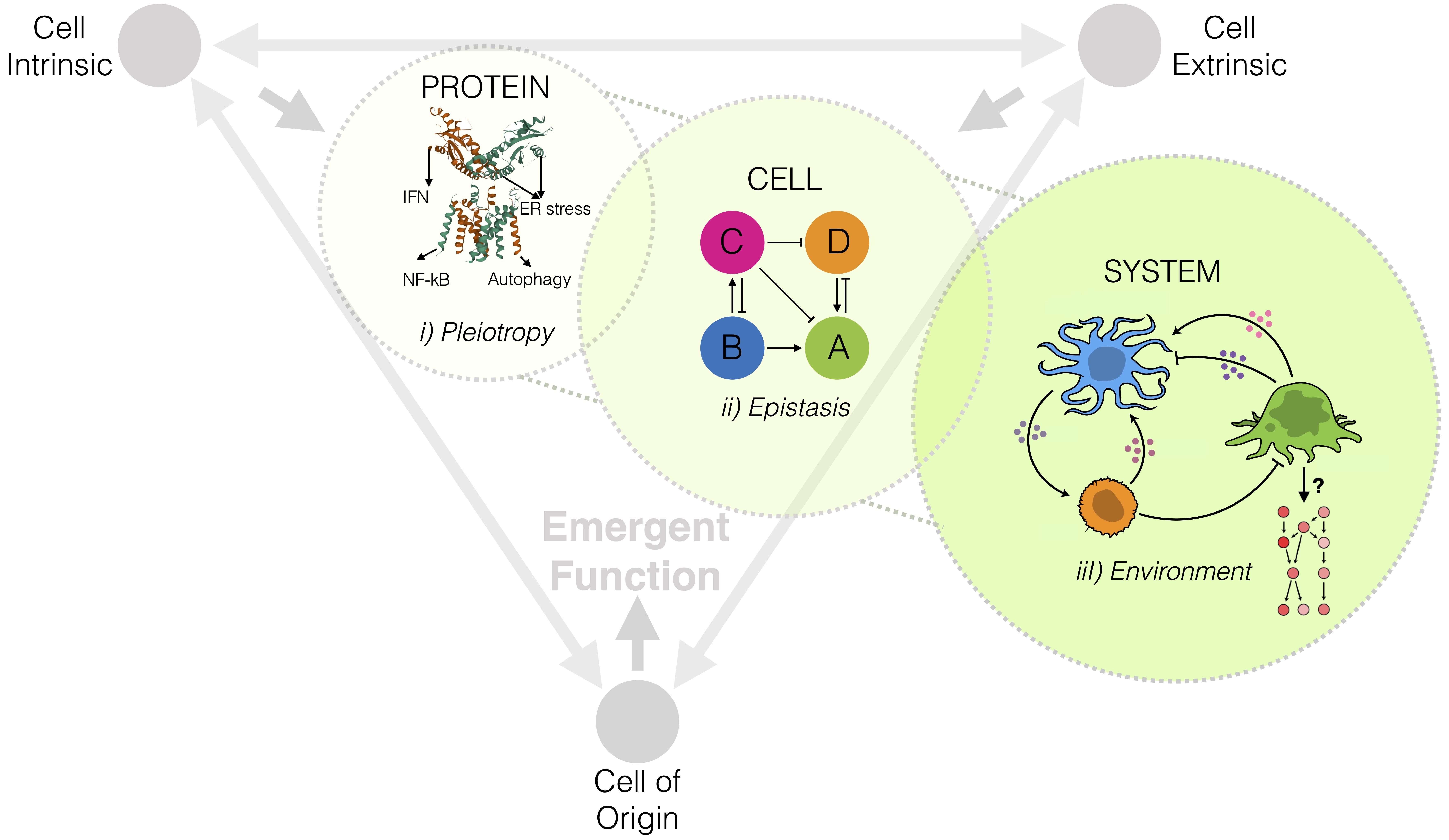
The genotype-to-phenotype problem. Determining how genome-encoded components perform cellular function is complicated by the pleiotropy found in biological systems. Pleiotropy refers to when a single gene influences multiple unrelated traits. As such, perturbations to a single gene can affect multiple, independent cellular responses – complicating our ability to infer sequence-function relationships. This problem, otherwise known as genotype-to-phenotype mapping, represents one of biology’s most intractable challenges. Combining high-throughput single cell sequencing with development of innovative computational tools, synthetic biology approaches and mouse models of cancer, our lab asks how the same genome-encoded components can give rise to diverse phenotypes, or cellular functions, at different stages of disease progression. We focus on cancer metastasis as our model of a complex, evolutionary process and develop novel approaches that tackle fundamental problems encountered in genotype-to-phenotype mapping, including: 1) Pleiotropy, 2) Epistasis and 3) Microenvironmental Crosstalk.
Figure 1. Tackling the genotype-to-phenotype problem in cancer evolution across scales.
One way in which cells adapt diverse functional responses is through context-dependent interactions with their microenvironment. Cells typically send messages to their microenvironment by emitting ligands, which bind to complimentary receptors on the surfaced of target cells; then triggering a change in the behavior of the target cell. Complimentary ligand-receptor-pairs are well annotated and methods have been developed to infer cell-cell interactions from single cell expression data based on their mutual expression. However, existing work views each cell as an independent unit. We have taken the alternative view that cells sampled from the same tumor microenvironment are fundamentally participating in an underlying biological process and are working towards generating a principled understanding of how cells coordinate their behavior; which has the potential unlock deep biological insights into cancer progression, and suggest new targets for effective treatments. Exploiting inter- and intra-sample variability, we are developing new methods to infer the effect of a ligand-receptor-mediated interaction on the tumor microenvironment and to characterize intercellular network dynamics.
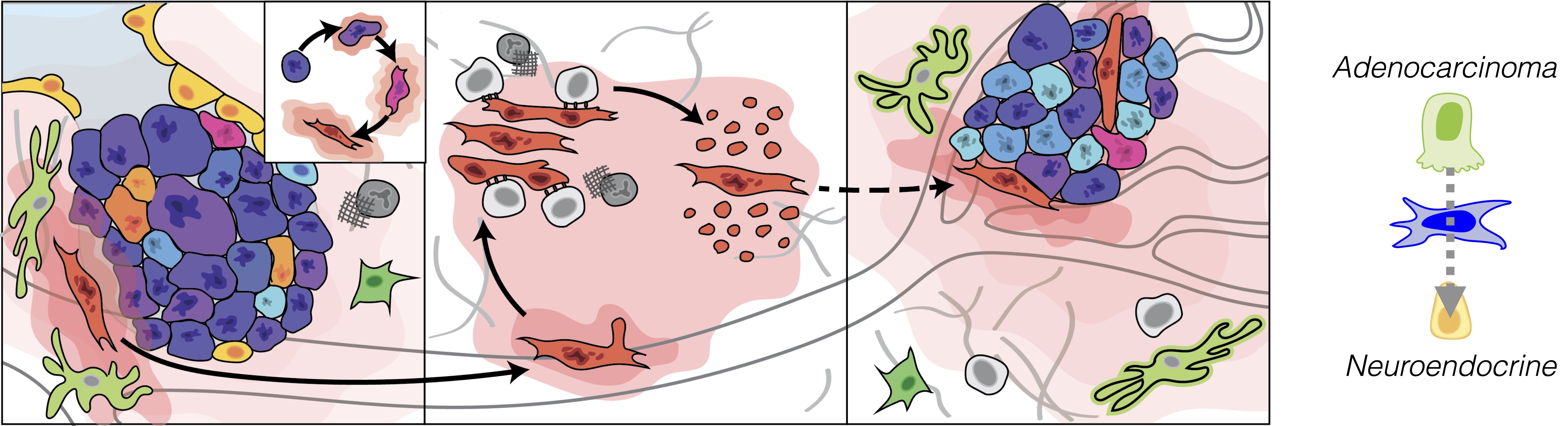
A focus on the cytosolic DNA sensing pathway. Most cancer deaths are caused by metastasis; an emergent phenotype that requires diverse functions including dissemination, adaptation to a less hospitable microenvironment, immune evasion and regeneration of a secondary tumor at a distant site. Chromosomal instability (CIN) is a key driver of this transition through chronic sensing of genomic double-stranded DNA (dsDNA) in the cytosol (Nature, 2018). In fact, it is one of the only conserved genomic drivers of cancer metastasis (Cell, 2022). This process triggers inflammatory signaling that regulates cellular microenvironment crosstalk (Nature, 2023), events central to a wound-healing response (British Journal of Cancer, 2021).
Figure 2. Emergent functions in cancer progression: metastasis (left) and histologic transformation (right).
Current Projects:
- Modeling cellular crosstalk in the tumor microenvironment.
- Chromosomal instability and the tumor ecosystem.
- The human brain metastatic niche.
- Cytosolic nucleic acid sensing pathways in cancer.
- STING functional elements in cancer progression.
- Lineage plasticity in lung cancer.
Bio
Laughney completed her undergraduate training in Physics at the University of Vermont and earned her PhD in Engineering at Dartmouth College in the lab of Brian Pogue, developing functional spectroscopy approaches. She then carried out her postdoctoral work in single cell imaging and genomic methods at Harvard Medical School and Memorial Sloan Kettering Cancer Center for applications in cancer biology. In 2019, she became an Assistant Professor and Member of the Meyer Cancer Center and Institute for Computational Biomedicine at Weill Cornell Medicine. Her lab now integrates single cell sequencing with innovative computational and synthetic biology tools to tackle the genotype-to-phenotype problem in evolving, multi-cellular processes like cancer metastasis.
Distinctions:
- Burroughs Wellcome Fund Career Award at the Scientific Interface
- Kellen Junior Faculty Award
- Regeneron Prize for Creative Innovation Finalist
- SLAS Innovation Prize Finalist
