
Research
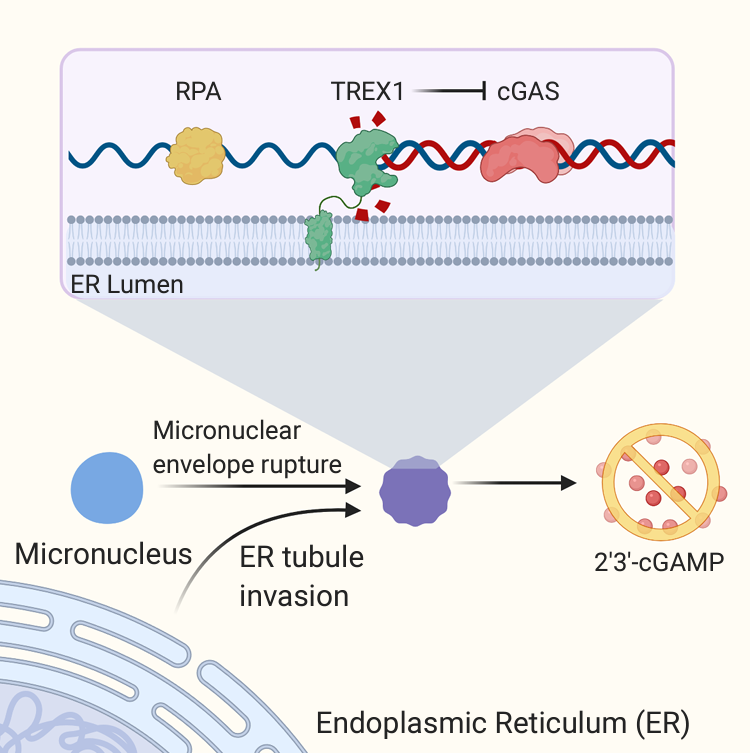
The Maciejowski lab investigates how innate immune responses, ordinarily linked to defense against pathogens, respond to cancer-intrinsic aberrations. The innate immune system relies on a limited number of germline-encoded receptors that recognize features associated with pathogens. Once activated innate immune receptors engage various cellular signaling pathways to activate wide-ranging, anti-pathogenic responses that include inflammation and production of other anti-viral or anti-microbial proteins. The distinction between pathogen and self can often be blurred especially within the cancer setting. One common feature associated with both pathogenic infection and cancer-associated genomic instability is the presence of double-stranded DNA in the cytosol. The cGAS-STING pathway senses and responds to cytosolic DNA by producing a wide-ranging pro-inflammatory response. The cGAS-STING pathway plays critical roles in anti-tumor immunity and the efficacy of immunotherapies. It is therefore critical to understand how the pathway is activated and regulated within the cancer setting. The Maciejowski lab discovered that the endoplasmic reticulum-associated TREX1 nuclease limits cGAS-STING activation in genomically unstable cancer cells by degrading DNA within aberrant nuclear compartments termed micronuclei, a major source of cytosolic DNA in many cancer cells (Figure 1). TREX1 activity at micronuclei depends on its associated with the endoplasmic reticulum. The lab now seeks to 1. Determine the mechanisms of ER-dependent TREX1 recruitment to MN, 2. Dissect the role of TREX1-interaction partners and non-catalytic elements of TREX1 in cGAS-STING regulation, 3. Pursue therapeutic opportunities arising from TREX1 micronuclei attack, 4. Understand how DNA damage at micronuclei impacts the cancer genome. Our long-term goals are to identify strategies to improve immunotherapies and obtain insights into cancer genome evolution.
Parallel work in the lab focuses on the role of the APOBEC3 family of cytosine deaminases in cancer mutagenesis. The APOBEC3s, which target ssDNA of viruses and retroelements as part of the innate immune defense, are proposed to cause two prominent mutational signatures. APOBEC3 mutations are among the most prevalent mutational signatures detected across human cancer and associate with aggressive disease features, such as metastasis and therapy resistance. It is therefore critical understand the fundamental mechanisms underlying APOBEC3 mutagenesis in cancer. The lab used cancer cell line models with active APOBEC3 mutagenesis to identify the APOBEC3A paralog as a major driver of mutagenesis indicating that APOBEC3A is a major enzymatic source of mutations in cancer (Figure 2). The current goals of the lab are to 1. Elucidate mechanisms underlying APOBEC3A-dependent mutagenesis; 2. Identify instigators of APOBEC3A dysfunction; 3. Dissect the roles of individual APOBEC3 enzymes in cancer mutagenesis.
Figure 1
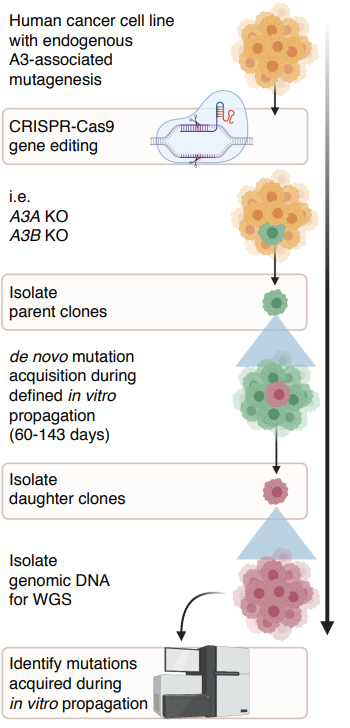
Figure 2
Current Projects:
- Chromosomal instability
- cGAS-STING
- APOBEC3 mutagenesis
- Cancer genome evolution
Bio
John Maciejowski, Ph.D., joined the Sloan Kettering Institute as an Assistant Member in the Molecular Biology Program in 2017. John performed his doctoral work at MSK as part of the inaugural class of the Gerstner Sloan Kettering Graduate School of Biomedical Sciences. While at GSK, John worked in the lab of Prasad Jallepalli studying the mechanisms that maintain high fidelity chromosome segregation during mitosis. Following his graduate work, John joined Titia de Lange’s lab at Rockefeller University, where he identified a link between telomere dysfunction and the genesis of complex clusters of chromosome rearrangement and hypermutation.
Distinctions:
- Pershing Square Sohn Prize
- Pew Biomedical Scholar
- V Foundation Scholar
- Pathway to Independence Award
- Regeneron Prize for Creative Innovation
- Tri-Institutional Breakout Prize
- Jane Coffin Childs Fellowship
