Research
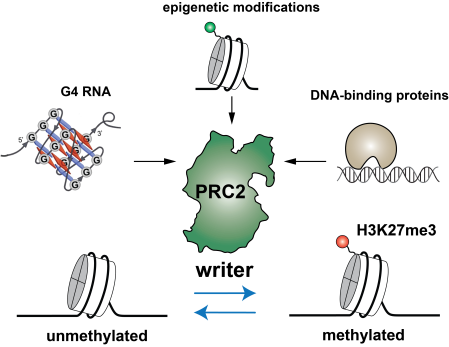
Body development requires precise gene expression in a spatial and temporal manner, which is controlled by epigenetics. H3K27me3 (trimethylation of lysine 27 on histone H3) is the hallmark for facultative heterochromatin, which dynamically regulate gene repression. During cardiac differentiation, H3K27me3 is deposited on pluripotency genes, erased on cardiac genes, and maintained on other developmental genes of ectopic lineages. The precise level of H3K27me3 across the genome is essential for the ON/OFF switch of gene expression, but it remained unclear how H3K27me3 is regulated in a coordinated temporal and spatial manner. We aim to understand how dynamic macromolecular interactions including protein-protein and protein-nucleic acid interactions regulates the specificity of the H3K27me3 “writer” and “eraser” enzymes. The H3K27me3 writer complex, Polycomb Repressive Complex 2 or PRC2, is regulated by DNA-binding proteins, G-quadruplex RNA and other chromatin modifications. Understanding the regulation mechanism will open to door to further identification of novel therapeutical targets to treat diseases.
Figure 1
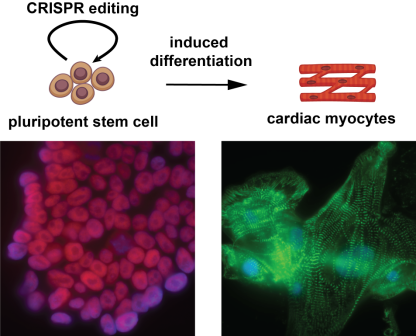
The advances in stem cell biology enable researchers to study how epigenetics regulate developmental process in a petri dish. We are studying cardiac differentiation using human induced pluripotent stem cells or embryonic stem cells. Recent advances showed that genes encoding epigenetic “writer” and “eraser” enzymes are frequently mutated in patient with congenital heart defects. However, treatment or early prevention methods are hampered by the lack of knowledge on how the epigenetic landscape is precisely regulated. Our lab uses stem cell cardiac differentiation models to study how these epigenetic enzymes and their disease mutations regulate gene expression during cell differentiation.
Figure 2
We are also interested in building innovative epigenome editing tools, which can be used for locus-specific manipulation of the epigenetic landscape. These tools are built on our mechanistic understanding of the epigenetic regulators such as noncoding RNAs and have the potential for correction of gene expression in diseases.
Current Projects:
- Epigenetics
- Nucleic acids
- Stem cell
- Cardiac differentiation
Bio
Yicheng obtained his bachelor’s degree in biological sciences from Tsinghua University in Beijing, and then he joined Jane Jackman's lab at Ohio State to pursue his PhD research on RNA repair and reverse RNA polymerization in Dictyostelium (a social amoeba) and budding yeast. After completion of his PhD study, he joined Tom Cech's lab at University of Colorado Boulder to study how RNA-protein interaction regulates epigenetic repression. Yicheng established his independent research lab at Weill Cornell in August 2021, and he is interested how molecular interactions among RNA, DNA and protein regulate gene expression in cell identity, development and diseases.
Distinctions:
- American Heart Association Career Development Award (2022)
- NIH Pathway to Independence Award (2020)
