
Anoushka Joglekar grew up in Pune, India, a metropolis with rich historical and cultural roots dating back several centuries. Joglekar came to the United States to pursue her undergraduate studies at the Ohio State University and joined the Tri-I Computational Biology and Medicine (Tri-I CBM) in 2017. Part of WCGS’s appeal was the direct access to some of the greatest research labs in the country. “I had the freedom to choose the focus of my research and I appreciate the highly collaborative environment of shared intellectual and physical resources,” Joglekar says. “And being able to live in NYC is a bonus.”
In the second year of her PhD training, Joglekar joined the Hagen Tilgner Lab in the neurogenetics department where they focus on alternative splicing and develop technology to investigate isoforms at a single cell level. “Alternative splicing is the process by which the same pre-mRNA molecule can be modified to produce multiple mRNA transcripts which are similar in structure,” explains Joglekar. “We refer to these protein precursors as mRNA isoforms because they come from the same gene, but encode distinct protein isoforms.”
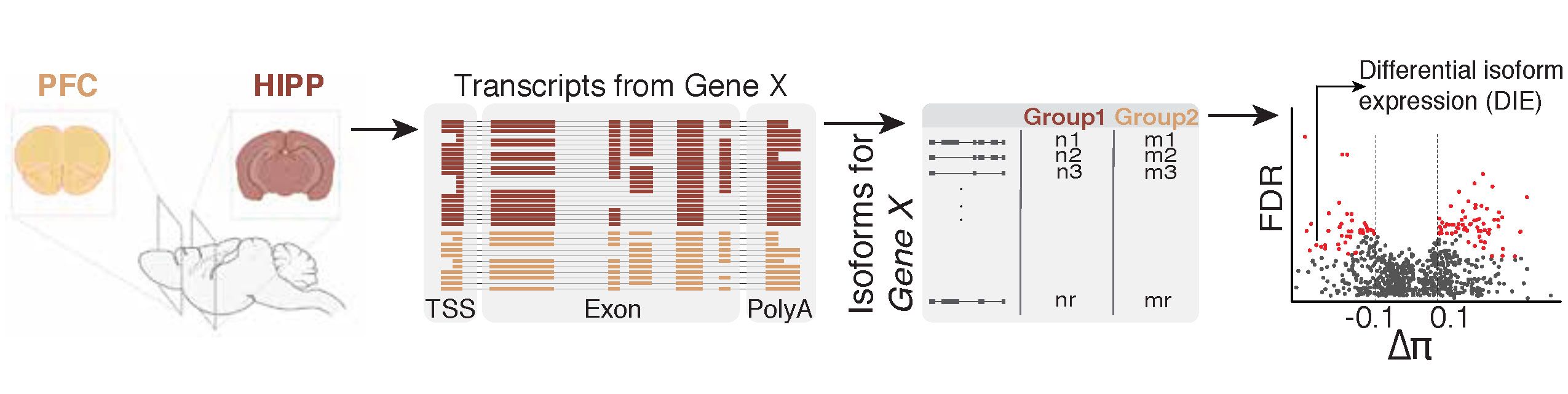
scisorseqr: a method to quantify differential isoform expression changes between any two conditions.
A few years ago, the Tilgner lab developed a technique where they could get the full-length transcript information from single cells. However, there were no existing methods for quantifying the differences in transcript usage, Joglekar clarifies, at least none that were efficient. Joglekar’s dissertation research focuses on developing computational tools to analyze data generated from the technology. A Harvey Lecture series that she attended caught her attention and inspired her research topic. “Dr. Adrian Krainer described a true bench-to-bedside story of developing the first antisense oligonucleotide therapy to treat Spino Muscular Atrophy, a lethal splicing-mediated disease,” says Joglekar. “The degree to which splicing is overlooked in the field of transcriptomics, especially in ‘genome-wide’ studies really struck me then.” The underappreciation in the field was not willful, but limited by technology. Joglekar remains enthusiastic and hopeful – “I started working on it as the tide was turning. There was an explosion in the field of single-cell transcriptomics which allows one to get signals from individual cells as opposed to an averaged-out bulk signal,” she says of her research. “This almost fortuitous timing of the technology development allowed us to ask the questions we were most interested in: do individual cells expressing the same genes also express the same isoforms?”
Recently, Joglekar’s work has resulted in a first author paper in Nature Communications entitled “A Spatially Resolved Brain Region- and Cell Type-specific Isoform Atlas of the Postnatal Mouse Brain.” In this study, the lab develops a method to measure differential splicing differences and apply it to two brain regions at a developmental time point. The findings show there are cases where the micro-environment can influence the splicing profile of the same cell-type in two different regions, indicating that splicing patterns are not just cell-type mediated, but are also brain-region mediated. “We take it a step further to show that this spatial separation is an important consideration by applying our methods to newer techniques that allow for the spatial encoding of transcripts,” Joglekar says, “additionally, isoforms have more information than gene counts and there is a spatial component to isoform expression that should not be ignored.” Furthermore, the discovery illustrates these techniques can be applied to study neurodegenerative diseases to holistically enquire about the role that individual cell types play in wreaking havoc in specific brain regions.
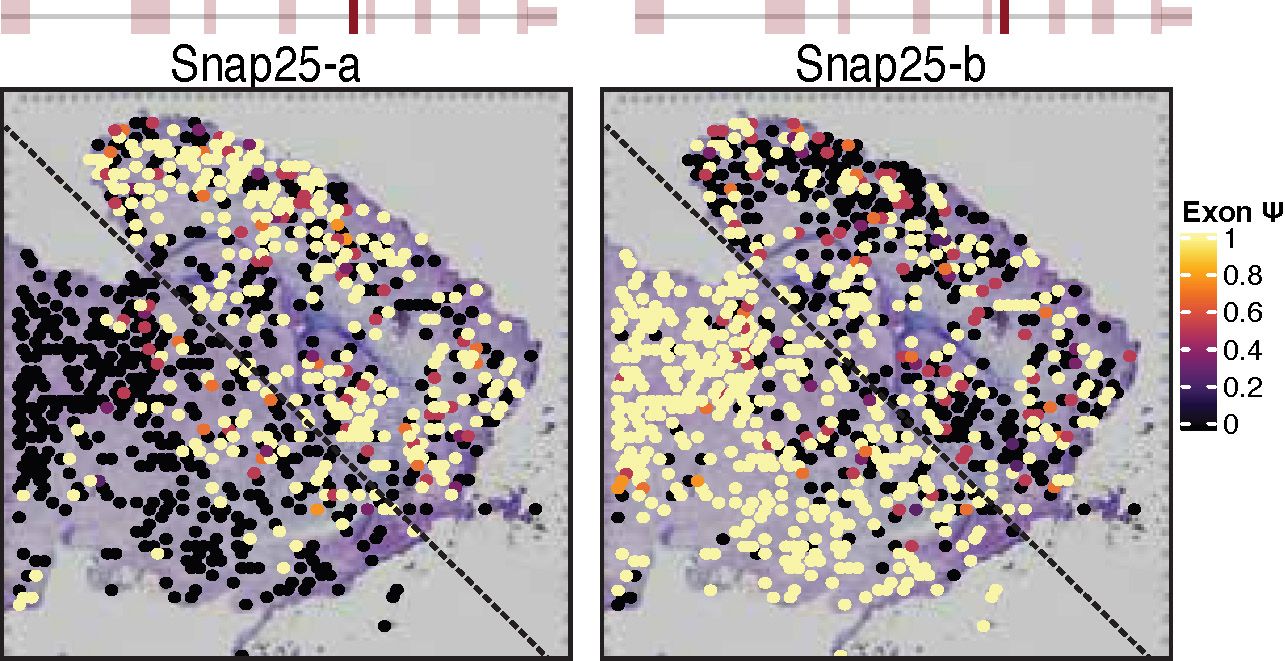
Long read sequencing on spatial transcriptomics technology allows for the spatial mapping of isoform expression. The developmental switch of Snap25 isoforms is seen as a gradient of isoform expression and is not uniform across all brain regions.
Joglekar’s research seeks to provide resources to biologists to query the splicing signatures of their genes of interest at different points in developmental time and space, such as the brain regions. In the long term, she hopes their approach will be the norm rather than the exception. “There is so much information right under our noses that can help fine-tune therapeutic targets,” says Joglekar. “I believe science is a team effort and there’s no point wasting resources and reinventing the wheel, which is also why we’ve developed and made our computational tool available for people to use to analyze their data.”
At WCGS, Joglekar is an active member in the Safe Space and Open Dialogue group of the Social Justice and Anti-Racism Task Force (SJAR). Joglekar used to think her voice and opinions did not matter since she does not identify within the historic norms of underrepresented minority groups. “In hindsight it’s a little silly,” she says. “However, for the past few years, I saw rising levels of injustice and silent compliance. I wanted to keep having important conversations with people who were not aware of the culture of toxicity and intolerance they had implicitly embraced.” Joglekar also believes that misunderstandings often arise from a lack of communication, and says “I want to be able to open the doors to frank dialogue as opposed to echo chambers of myopic perspectives.”
As for her plans after graduation, Joglekar says she is leaving her options open. “The immediacy of impact in industrial research is just as appealing as the freedom of pursuit in academia, so I’m going to hold off on making the decision for a bit longer.” Pre-pandemic, Joglekar used to do aerial arts, brunched with friends, and went to shows. Over the past year, she has reconnected with her passion for books, watching TV shows and spending time in the kitchen exploring different recipes.
