
Research
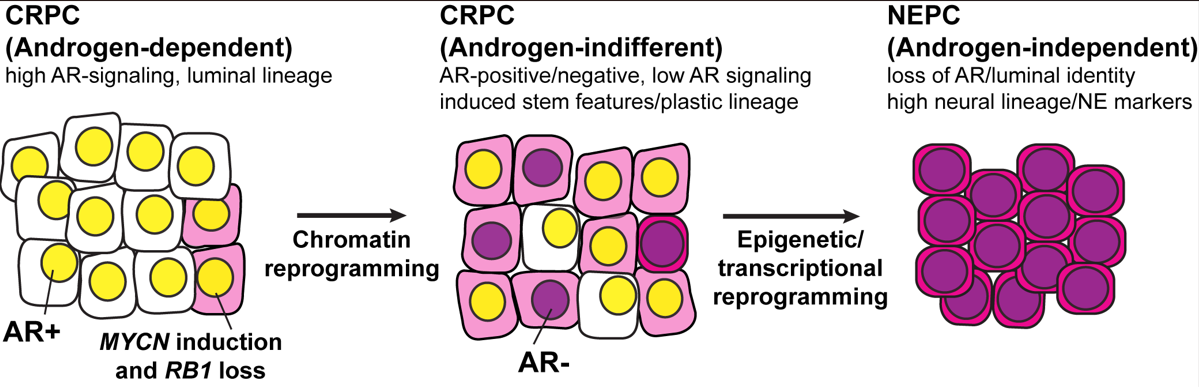
Prostate cancer arises as an androgen driven disease and therefore androgen receptor (AR) targeting therapies have been a major focus of prostate cancer treatment. Lineage plasticity, a process by which differentiated cells lose their identity and acquire alternative lineage programs, has recently been identified as a mechanism of resistance to targeted therapies in several cancer types including prostate cancer. For prostate cancer, this plasticity can manifest as histologic transformation from an AR-positive castration resistant prostate adenocarcinoma (CRPC) to an AR-indifferent, small cell carcinoma that expresses neuroendocrine markers, often referred to as neuroendocrine prostate cancer (NEPC, Figure 1). NEPC is clinically aggressive, frequently metastasizes to visceral organs, and carries a poor prognosis. The development of effective treatment for patients with NEPC remains an unmet clinical need. A thorough molecular understanding of lineage plasticity is needed for the development of strategies to treat, prevent, or reverse NEPC progression. Although NEPC tumors arise clonally from CRPC and share genomic alterations, there is significant epigenetic deregulation during the transformation process. However, mechanistically, we still do not know how these epigenetic alterations arise and how best to leverage these alterations as a therapeutic opportunity. Our data from patients and in vivo (e.g. genetically engineered mouse model and PDXs), in vitro and ex vivo models (patient-derived organoids) suggest that induction of specific transcription factors (e.g. N-Myc, E2Fs) and pioneer factors (e.g. FOXA1/2) results in a reprogramming of chromatin accessibility that is androgen-dependent and that drives epithelial plasticity and the acquisition of clinically-relevant, NEPC molecular program. Our data also suggests that this progression involves a reprogramming of the epigenome (e.g. EZH2 activity and DNA methylation). Interestingly, we found that specific molecular or pharmacological interventions can revert the NEPC phenotype to a luminal—more clinically manageable—adenocarcinoma phenotype. In summary, the over-arching aim of the Rickman Lab is to characterize the interplay between specific transcription factors, pioneer factors and epigenetic modifiers in driving progression of prostate cancer to a lethal form of the disease and how best to target this progression.
Figure 1
Current Projects:
- Transcription and pioneer factors driving lineage plasticity
- DNA methylation reprogramming
- Crosstalk between DNA methylation and EZH2 activity
Bio
Dr. Rickman earned his Ph. D. in molecular biology from Mt. Sinai School of Medicine, New York, New York in 1997. He then trained as a postdoctoral fellow with Dr. Samir Hanash at the University of Michigan Medical School from 1998 to 2001. He was then recruited to be project leader for the French Cancer Biomarker Program initiated by the (Ligue Nationale Contre le Cancer in Paris, France. Dr. Rickman then joined the faculty at Weill Cornell Medicine as an Instructor in 2007, Assistant Professor in 2009 and then Associate Professor in the Department of Pathology/Laboratory Medicine and the Cell and Developmental Biology Program in 2017.
Distinctions:
- New Investigator Award, Department of Defense, 2008
- Prostate Cancer Foundation Challenge Award, 2014
- IMPACT Award, Department of Defense, 2017
